

The discipline of metabolomics is the analysis of the metabolome: the small-molecule complement of a biological system. Biological small molecules (metabolites) are heterogeneous and diverse. They underpin the biochemical reactions that give rise to living systems, converting glucose into energy, sending and receiving information in the form of chemical signals and providing the building blocks for complex macromolecules.
While measurement of a single or a handful of metabolites has provided insights into health and disease for many years, the advent of metabolomics allows us to analyze a large portion of the whole metabolome simultaneously, giving information not only on specific metabolites of interest, but also an unbiased view of the complex changes and interactions that take place in functioning biochemical networks.
Lipidomics
Lipidomics is concerned with the characterisation of lipids, a diverse group of molecules found in all biological organisms that have pivotal structural, signalling and metabolic functions. We bring expertise in the identification and quantification of a wide variety of lipids from biological and clinical samples. We employ mass spectrometry in both global and targeted lipidomic analyses to investigate the key role that lipids play in human, animal, plant and microbial systems.
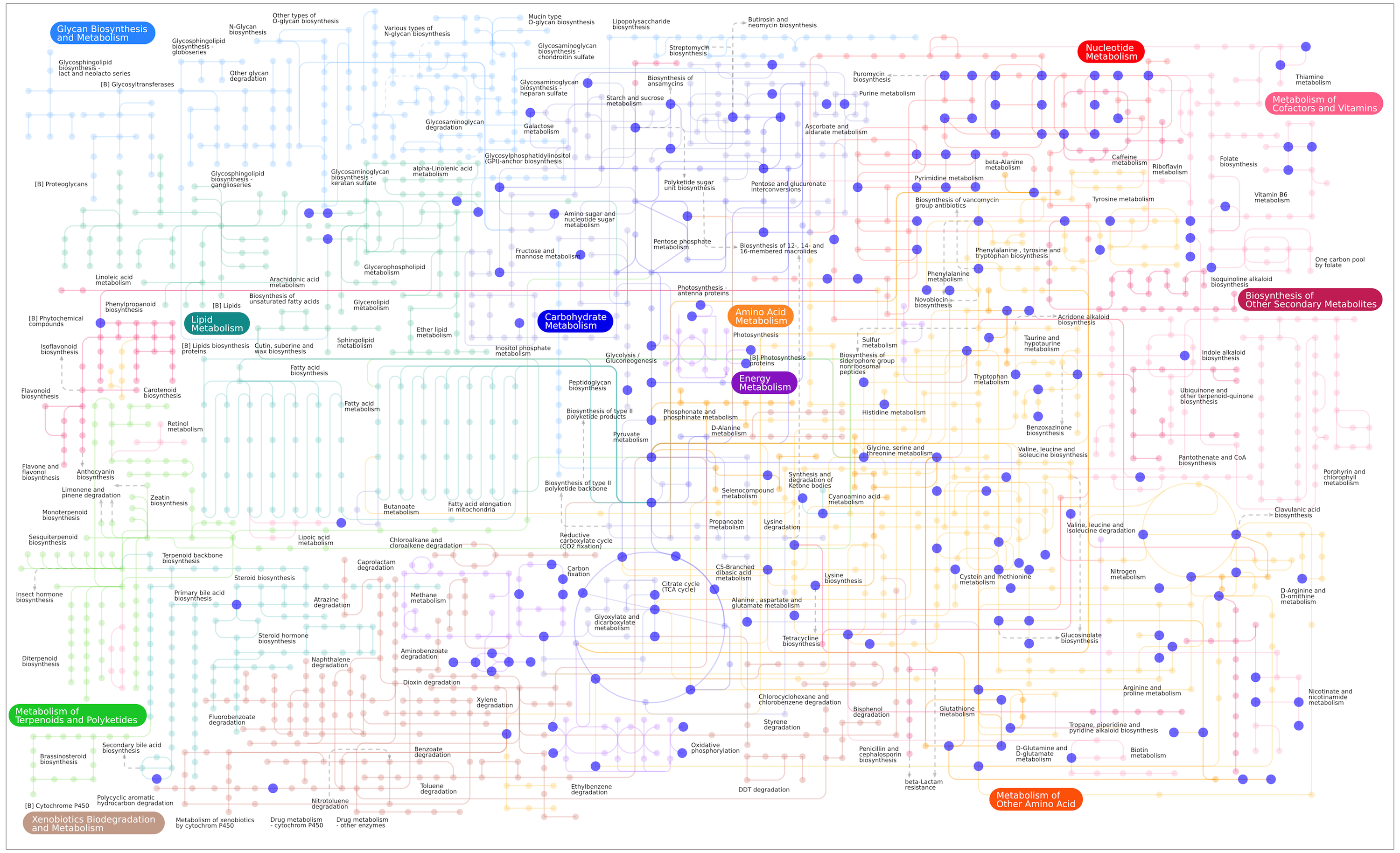
Our facility routinely runs over 150 reference compounds per analysis, covering a range of common metabolic pathways as illustrated in our Pathway Map of Standards.
The metabolome can be measured in virtually any biological system. Application areas vary widely, from drug mode-of-action to the effects of climate change.
Types of Projects
We can carry out various types of analysis, some more routine than others. If you do not see the method of interest, please contact us and we will assess its feasibility and discuss options with you.
In general the most common types of experiment are:
Preparing SamplesSending SamplesAnalysing SamplesOther
No. We only accept samples that have already been extracted.
There are two reasons for this. Firstly, we are unable to support the array of ribolysers, sonicators, French presses, etc that would be used for lysis of the wide variety of organisms that we analyse. Secondly, the quicker the samples are extracted into solvent, the closer the samples will be to the in-vivo metabolic state. Samples should be extracted as quickly as possible after sampling; if that’s not possible then please be consistent. For example, it is better to take 5 minutes for each sample than to vary extraction time between 10 seconds and 5 minutes.
We can provide a protocol for extracting metabolites from different organisms/tissues. For cells, we recommend starting from a ~5µL pellet of cells. For fluids, we’d prefer ~5–100µL depending on the concentration of the liquid (i.e. ~100µL of CSF or serum, whilst 5µL of cell culture medium is more than sufficient).
We recommend a minimum of three biological replicates per sample. This should be increased to 6 or more replicates where metabolomics is carried out on whole multicellular organisms, depending on the inherent variation in your system. Very carefully controlled samples (e.g. cell culture) need less replicates than highly variable ones (e.g. canine saliva).
Yes. The protocol for carrying out extraction from cells includes a step for analysing supernatant. Briefly, you should take 5μL of your supernatant and add 200μL chloroform/methanol/water (1:3:1 ratio) to it. Mix by vortex and centrifuge, then supply to us.
If they are eukaryotic cells without a tough cell wall, you can simply add an appropriate amount of extraction solvent (about 400μL for a 24-well plate, for example), and leave them on a rocker at 4 ̊C for 1 hour. You can then aspirate the supernatant, vortex and spin down, and then supply to us.
Please contact our Facility Administrator to log your project and receive the appropriate sample and project forms. We will run a batch of samples only if the samples and the sample list are matched. Please label all sample vials or plates clearly, using indelible ink.
Samples should be shipped on dry ice. Cryotubes would be ideal for transportation as they are screw-capped and therefore less likely to leak, sealing with parafilm can also be helpful. We will only accept samples that are supplied in a comprehensively labelled sample box (e.g. 15cm x 15cm) — this not only makes them easier to store for us, but also ensures they neither get mixed up in transit nor become difficult to find when we receive them. Please also label each tube clearly. When you fill in the sample submission form, please use the same labels as are on the tubes. For longer journeys especially, we would also suggest wrapping parafilm around the screw-top vials and enveloping the entire box in plastic; the samples should then be buried in the middle of the dry ice.
Before sending your samples please fill in the sample submission and project forms; availabile from our Facility Administrator. This ensures that we have all the required information about your sample, including the analysis plan, and also makes sure that we are able to run the samples in a random order. It is important that the labelling on the sample tubes matches the labelling on the submitted sample file. Internationally shipped samples should be supplied in screw-capped tubes, to avoid leakage during transit.
Please note that it is the responsibility of the user to ensure that the samples arrive at the facility. We would advise users to track the progress of their package daily.
Our postal address is:
IPA - Mass Spectrometry
Room B4.10, Joseph Black Building
College of Medical, Veterinary & Life Sciences
University of Glasgow,
G12 8QQ
UK
Please enclose a printed copy of your sample submission form with your samples, and alert our Facility Administrator to let us know that the samples are on their way.
Important: Please follow the correct procedure given above for packaging the samples.
University of Glasgow users can deliver the samples to the Metabolomics Laboratory using the internal mail system. It is not advised to send samples on a Friday afternoon as they might not reach the facility until the following Monday morning.
To prioritize the delivery of your samples, please mark the box clearly to indicate that it contains dry ice. Please enclose a printed copy of your sample submission form with your samples, and alert our Facility Administrator to let us know that the samples are on their way.
Yes. If you are delivering your samples in person then please confirm a delivery time with a member of staff before travelling to the laboratory. As the building reception is manned only from 9:00–15:30, please do not attempt deliveries outside these times.
We can analyse virtually any biological fluid, or tissues from which fluid can be extracted (including cells). For example, this includes serum, blood, urine, CSF, and synovial fluid.
For liquid chromatography (LC)–MS-based metabolomics, we are equipped with 3 Thermo Orbitrap instruments: an Exactive, a Q-Exactive, and an Orbitrap Elite. A Trace Ultra Gas chromatography system coupled to an ITQ 900 Mass Spectrometer (GC–MS) is also available for volatile compounds that escape detection using the LC systems.
In general, if you're looking for depth of coverage of the metabolome, we recommend LC–MS as several hundred metabolites can be detected from a single experiment. In contrast, 1H NMR is likely to be limited to detecting 50–100 metabolites. The principal benefit of NMR over LC–MS is the reproducibility, throughput, and absolute quantitation available. LC–MS can only provide relative quantitation (e.g. that there is 10x more arginine in sample A compared to sample B) unless an isotopically labelled standard can be provided by the user, preferably spiked in during sample preparation at known concentrations.
We use a ZIC-pHILIC platform for our analysis, so the majority of compounds that we detect are polar. However, we detect a substantial number of lipids in the wash-through and can assign putative identifications to them using our software. Please contact us for a list of metabolites that are routinely detected.
Our instrumentation detects in both positive and negative ionisation modes, and, on average, 500–1,500 putative metabolites are detected per run. Approximately 250 of these will be validated against our panel of standards and are publishable directly according to the Metabolomics Standards Initiative. The remainder will require separate validation, which we can also provide.
Obtaining an absolute abundance of a metabolite requires the use of a deuterated standard. Due to the complexity of the metabolome, we are unable to provide deuterated standards for the thousands of compounds in metabolic systems, and therefore we can only provide relative abundances. However, once you have identified a particular metabolite of interest, we can validate and perform absolute quantitation if an isotopically labelled standard is available.
Our analyses are carried out using the openscource software PMP.
We use a minimum of 50μL of sample. Mass spectrometry is a complex technique and this volume allows for limited troubleshooting. For further analyses such as validation, additional sample is required, so please provide a minimum of 200μL unless you’re really sample limited.
Waiting times range from one month to six weeks depending on the length of the queue. We will give you an estimate of the waiting time when your samples have been submitted.
Yes. Please let us know at the outset whether this is your preference.
Only in very specific circumstances. In order to provide a high quality facility, it is essential that we limit access to the instrumentation to a handful of expert users.
Only in very specific circumstances. In order to provide a high quality facility, it is essential that we limit access to the instrumentation to a handful of expert users.
While our instrumentation is capable of generally providing an accurate chemical formula, many isomers are present in biological systems, and these are impossible to distinguish even by accurate mass. To report a hit as an 'identification', it must be compared to an authentic standard, of which we run about 250 with each batch. For other, less commonly seen compounds, we often have to buy in a standard and confirm the match to the compound in your sample by chromatographic retention time, mass and fragment pattern. We provide this as a costed resource in addition to the standard analysis.
The cost per sample depends on the number of samples you are submitting and the amount of bioinformatics you require. Please contact us to discuss your requirements, and we can provide you with a quote.
The cost per sample includes positive and negative ionisation analysis on our Orbitrap instrument as well as data analysis. Our data analysis product consists of a comparison of metabolites that are detected between the samples. This spreadsheet will contain: the list of putative metabolites found in the samples; their biochemical annotation, and their relative abundance between the groups of samples. You can also have a meeting or teleconference with the bioinformatician to discuss your data.
Note that we are unable to provide a thorough biological interpretation of the metabolite dataset: this is best done by the user, who will be more familiar with the biological context of the system of interest. However, if more support is required, a session can be booked with a bionformatician, who will help you to navigate your dataset.